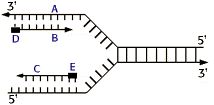
2) Examine the following diagram of a replication fork:

a) The strand labeled B is the______________.
b) The newly synthesized DNA strand labeled C is the_____________.
c) The black boxes labeled D and E are______________.
3) The entire genome of the fruit fly D.melanogaster
consists of 1.65 x 108 bp. If replication at a single
replication fork occurs at the rate of of 30 bp/sec calculate the minimum
time required to replicate the enire genome if replication were inititaed:
a) at a single bidirectional origin.4) Why are two different DNA polymerase enzymes required to replicate the E.coli chromosome?
b) at 2000 bidirectional origins.
c) In the early embryo, replication can require as little as 5 minutes. What is the minimum number of origins necessary to account for this replication rate?
5) Why do cells exposed to visible light following irradiation with ultraviolet light have a greater survival rate than cells kept in the dark after exposure to ultraviolet light?
6) a) Why does the addition of SSB (single stranded binding protein)
to sequencing reactions often increase the yield of DNA sequenced?
b) What is the advantage of carrying out sequencing reactions at 65
using DNA polymerase isoltaed from bacteria groing at a higher temperature?
7) A Sanger didexy sequence analysis was performed on wild type and a mutant gene. The sequencing gels are represented below. From these results
a) determine each sequence in full in the template starnd in the 5'->3' direction.WT MUTANT
b) determine the mutant site and the nature of the mutaional event.
___
___
___
___
___
___
___
___
___
___
___
___
___
___
___
___
___
___
___
___
___
___
___
___
___
___
___
___
______________________________________________________________________________
8) Under conditions where methionine must be the first amino acid,
what protein would be coded for by the following mRNA?
5'-CCUCAUAUGCGCCAUUAUAAGUGACACACA-3'9) A messenger acid is 336 nucleotides long, including the initiator and termination codons. How many amino acids are in the protein translated from this mRNA?
10) Your lab mate has designed a PCR experiment identical to the one shown in Figure 23.17 of your text, except that by mistake they used two oligonucleotide primers that were complimentary to the ones depicted in the figure (i.e.they hybridize to the correct correct site but on the opposite strand). Will PCR still amplify the correct product. Why or why not?
11) The recognition sites for the restriction endonucleases BglII and BamH1 are shown below (- represents the cleavage site).
BglII A-GATCT BamH1 G-GATCC
Why is it possible to construct recombinant molecules by combing target DNA cut with BglII and a vector cut with BamH1?
12) A PstI restriction fragment of DNA is mixed with pBR322 DNA (see Figure 23.3) that has been cleaved with PstI. The mixture is treated with DNA ligase and used to transform a mixture of E.coli cells. An aliquot of the culture is spread on a petri dish containing tetracycline. Which cells form colonies? How can colonies containing recombinant plasmids be identified?
Problem #1